- Assistant Professor
- Biological Sciences
- Natural Sciences Building S-239
- Email: gillisw @ oldwestbury.edu
- Phone: (516) 876-2746
Research Interests
- Comparative Developmental Biology
- Bioinformatics and Comparative Genomics
- Molecular and Cell Biology
"It is not birth, marriage, or death, but gastrulation which is truly the most important time in your life." - Lewis Wolpert
We truly live at an amazing time in biology. When I was still in my undergraduate studies, the first human genome sequence was released. What we found was a little startling, as our genomes are remarkably similar to very distantly related invertebrates such as flys or worms. This means at some point, both flies and humans shared an ancestor with a fairly sophisticated set of body patterning genes, with our differences in bodies (e.g. wings verus limbs) are largely the result of reuse and redeployment of these genes.
I’ve spent my career exploring many of the questions that this remarkable conservation brings up. This work has had me study how similar genes are used to pattern a diversity of animals, from marine worms, sea anemones, lancelets, and now to frog and mammalian cells systems. As a new assistant professor at SUNY SUNY Old Westbury I use frog embryos and mammalian cell culture to better understand key signaling pathways which normally control development, and whose misregulation lead to diseases such as cancer. Before coming to Westbury I was a NY-CAPS postdoctoral fellow at Stony Brook University, an NIH-IRACDA program at geared to train the next generation of researchers and teachers. For more info see my Promo Video made while a NY-CAPS Postdoctoral fellow https://www.youtube.com/watch?v=q5dqcgFnkOI
Selected Publications
- Strigamia maritima sequencing and annotation consortium (105 authors, including W.Q.Gillis (#30)). Prototypical Arthropod Gene Content and Genome Organisation in the Centipede Strigamia maritima. PLoS Biology (in press)
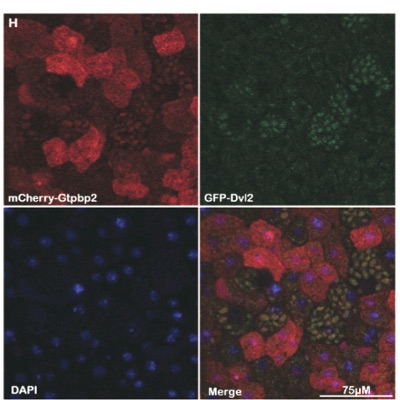
- Kirmizitas A, Gillis WQ, Zhu H, Thomsen GH (2014) “Gtpbp2 is required for BMP signalling and mesoderm patterning in Xenopus embryos”. Dev Biol pii: 2014 Aug 15;392(2):358-67
- Sorrentino GM, Gillis WQ, Oomen-Hajagos J, Thomsen GH “Conservation and Evolutionary Divergence in the Activity of Receptor-Regulated Smads” Evodevo. 2012 Oct 1;3(1):22
- Gillis WQ, St John J, Bowerman B, Schneider SQ “Whole genome duplications and expansion of the vertebrate GATA transcription factor gene family” BMC Evolutionary Biology, 2009, 9:207
- Gillis WQ, Bowerman B, Schneider SQ “The evolution of protostome GATA factors: molecular phylogenetics, synteny, and intron/exon structure reveal orthologous relationships” BMC Evolutionary Biology, 2008, 8:122
- Gillis WJ, Bowerman B, Schneider SQ “Ectoderm- and endomesoderm-specific GATA transcription factors in the marine annelid Platynereis dumerilii” Evolution and Development 9 (1): 39-50 Jan-Feb 2007
- S. Khan, R. Makkena, F. McGeary, K. Decker, W. Gillis, and C.J. Schmidt. “A Multi-Agent System for the Quantitative Simulation of Biological Networks”. In Proceedings of the International Conference on Autonomous Agents and Multi-Agent Systems, 2003.
- S. Khan, K. Decker, W. Gillis, and C.J. Schmidt. A Multi-Agent System-driven AI Planning Approach to Biological Pathway Discovery. In Proceedings of the International Conference on Automated Planning, 2003.
Presentations
- William Gillis (2013, March) “A large GTPase required for embryonic Wnt signaling through regulation of Axin stability” Talk presented at 2013 NWDB meeting, Friday Harbor, Washington
- Gillis W, Kirmizitas A, Ki D., Thomsen G. (2010, December) “Regulation of TGF-Beta Signaling and Embryonic Development by Large GTPases”. Poster presented at 50th Annual Meeting of the American Society for Cell Biology, Philadelphia, Pennsylvania
- Gillis W., Bowerman B.A., Schneider S.Q. (2008, March) “GATA Transcription factor evolution: Using genomic features to infer phylogeny” Talk Presented at the Northwest Regional meeting of the Society of Developmental Biologists, Friday Harbor, Washington.
- Gillis W.J., Bowerman B.A., Schneider S.Q. (2007, June) “The evolution of metazoan GATA factors”. Institute seminar given at EMBL, Heidelberg, Germany
- Gillis W.J., Bowerman B.A., Schneider S.Q. (2006, March) “Expression domains of two highly conserved GATA factors in Platynereis dumerilii elucidate an ancestral bilaterian split between neuroectodermal and mesodermal subfunctions” Talk presented at the Northwest Regional meeting of the Society of Developmental Biologists, Friday Harbor, Washington.
- Gillis W.J., Bowerman B.A., Schneider S.Q. (2005, July) The GATA family of Transcription Factors within the polychaete Platyneries dumerilii. Poster Presented at the annual meeting of the Society of Developmental Biologists, San Francisco, California.
- Gillis W.J., Bowerman B.A., Schneider S.Q. (2005, March) “The GATA family of transcription factors within the polychaete Platyneries dumerilii.” Poster presented at the Northwest Regional meeting of the Society of Developmental Biologists, Friday Harbor, Washington.
Lab members
- Anthony Lombardo
- Jonathon Doyle